Hossein Khedmat1,2 , Ali Karami2,3, Zahra Safiri2,3, Mohsen Amini1,2, Ali Bakhtiari2, Ashraf Karbasi1,2, Mojgan, Jayhounian2,4, Hamidreza Jalalian5, Saeed Taheri6
Baqiyatallah Research Center for Gastroenterology & Liver diseases1 &Medical Sciences2,
Biology Research Center3,
Department of Pathology4 & Medicine5,
Dr Taheri Medical Research Group6,
Tehran, Iran
Corresponding Author:
Dr. Hossein Khedmat
Email: Khedmat.h@gmail.com
DOI:
http://dx.doi.org/
48uep6bbphidvals|326 48uep6bbph|2000F98CTab_Articles|Fulltext Precise detection of H. pylori infection is relevant for clinical practice as well as for research purposes, and several invasive and noninvasive tests are currently available for diagnosis of H. pylori. Culture has long been the method of choice to detect infectious agents. However, culture has some limitation in a prompt detection as H.pylori is a very slow growing bacteria.[1] The sensitivity and specificity of different diagnostic tests for H. pylori detection varies widely.[2] Additionally, serology cannot detect the clearance of H. pylori and urease assays can lead to non-specific results due to the presence of other urease-positive bacteria and false negative results have also been reported in individuals taking proton pump inhibitors.[3,4] Methods based on molecular biology are considered highly specific and sensitive tests, and many PCR-based assays have been developed to detect H. pylori DNA in gastric biopsies, saliva and stool samples.[5,6] However, this technique is able to detect specific fragments but not viable bacteria, and its sensitivity also depends on several factors.[6]
Recently, we established a PCR based method for detection of Helicobacter pylori at our institution. The present study was therefore aimed to determine the sensitivity and specificity of PCR primers to diagnose Helicobacter pylori infection.
Methods
Patients
All patients who attended the outpatient clinic of gastroenterology with dyspepsia and underwent a diagnostic endoscopy evaluation with biopsy from March 2005 to March 2006 were consecutively included into this analysis. A total of 664 specimens from 166 patients (four biopsies each) were assessed. Biopsies were taken from the antrum of the patients for rapid urease test, histo-pathological examinations, and DNA analysis. Three specimens were sent for rapid urease test and histopathology and the remaining specimen was frozen for PCR analysis.
As culture of H. pylori from biopsy specimens was not performed in our study, any sample positive on histological examination as well as rapid urease test was considered as the gold standard for determination of the sensitivity and specificity of the PCR methods.
Histological examination
Paraffin-embedded tissue sections were stained with hematoxylin and eosin and the severity of gastritis was graded, according to the Sydney system.[7] Giemsa stain was used to to detect H. pylori.
Rapid-urease test
One antrum biopsy specimen was introduced with a sterile needle into a semisolid 2% urea agar and incubated at room temperature. Results were recorded up to 4 h after inoculation.[8]
Preparation of samples for PCR amplification
Genomic DNAs were extracted from all strains by method of Marais et al.[9] The extracted DNAs were dissolved in water, and solutions were prepared and used throughout the study. Briefly, the biopsy samples were ground and centrifuged for 5 min at 10,000×g. After the supernatants were discarded, biopsy specimens were resuspended in extraction buffer (20 mmol/L Tris- HCl, pH 8.0; 0.5% Tween 20) and proteinase K (0.5 mg/mL final concentration). The mixture was incubated at 56 °C for one hour after which the enzyme was inactivated by boiling for 10 min.
Using our nested assay, we were able to detect H. pylorispecific sequences at an estimated concentration of 20 picomoles. Five µL of DNA was used as the template for each PCR. Each sample was examined by four different PCRs. Primers used in this study were from, 16S rRNA (bp:521), Urease A (bp:411), Cag A (bp: 400), 26kDa ( bp: 303). The primer sequences and PCR conditions are listed in Table 1.

Statistical analysis
SPSS software (Statistical Product and Services Solutions, version 13.0, SPSS Inc, Chicago, IL, USA) was used to analyze the data. Statistical differences between patients’ subgroups were assessed using the chi-square test, the Fisher exact test for proportions, and the t test for continuous data. Values for P less than .05 were considered statistically significant.
Results
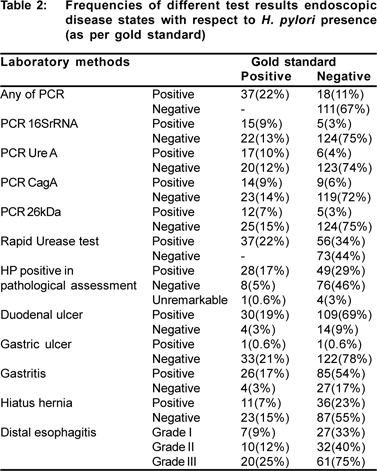
Complete data for this study was obtained on 166 patients (78 males, 88 females; age: 41.7±15.7 years). Of these, a total of 55 (33%) patients gave positive results for at least one of the PCR methods employed and by any methods (including PCR), respectively. Thirty seven samples (22%) were positive both by rapid urease test and by nested PCR. Eighty nine biopsies were positive by histologic staining (48.7%), of which 49 gave negative results by PCR.
Patient samples were considered to be positive for H. pylori by PCR amplification if any of the 521bp, 411bp, 400bp, 303bp bands was seen in the reaction. Of the 55 PCR positive samples, 23 (41.8%) showed 400bp band, 31 (56.4%) represented 303bp band, 20 (36.4%) revealed 521bp band, and 23 (41.8) presented 411bp band.
32 (19%) samples were positive by both PCR and pathological assessment, while 49 (29.5%) of pathologically positive subjects were negative for H. pylori DNA by PCR methods and 23 (13.9%) of PCR positive samples were negative by pathology. Nine samples were negative by pathology while positive by both rapid urease test and PCR methods; 41 cases were negative for PCR evaluations while they were positive by both rapid urease test and pathology; as well, only 4 cases who were positive by both PCR and pathology were negative by rapid urease test.
The degree of gastritis based on pathological evaluations using Sydney system was not associated with a positive result on PCR for H.pylori (p>0.1). However, patients with a positive rapid urease test had significantly higher grade of gastritis (Grade of Gastritis mean±SD: 2.3±1.3 vs. 0.3±0.8; respectively; p<0.0001).
Based on the gold standard, 37 of the patients examined as part of this study (22.3%) were diagnosed as H. pylori infected. Table 2 shows different variables of the study with respect to the study’s gold standard result. Computed sensitivity for 521bp, 411bp, 400bp, 303bp bands for detection of H. Pylori and exclusion of negative cases in this study were 41%, 46%, 38%, and 32%, respectively; the attributed specificity for each of the PCR methods were 85%, 86%, 84%, and 83%, respectively (Table 3).

Discussion
Appropriate diagnostic tools for diagnosis of a probable H. pylori infection in patients with dyspepsia is of utmost importance for physicians as well as for patients. The rapid urease test is the most frequently used diagnostic test for the diagnosis of H. pylori infection in patients with chronic gastritis in gastroenterology clinics. This method of diagnosis has several advantages including giving a prompt result for H. pylori infection even before the patient leaves the clinic, high diagnostic accuracy as well as its low economical strain on both patients and the health systems.[10,11,12] Histological diagnosis of H. pylori infection is the reserved method especially for patients with a negative rapid urease test and a high suspicion of infection or for exclusion of malignancy.
The general concept is that PCR is the most sensitive technique for the detection of microorganisms such as H. pylori. The detection of H. pylori in gastric biopsy samples by PCR has been assessed by several researchers representing high sensitivity and specificity usually over 95% as compared to other invasive methods.[13,14,15,16]
In this study, we examined the strength of PCR methods in the detection of four H. pylori recognized alleles. We found that PCR has a very low power to detect H.pylori infection among our patients. There may be several reasons for this; first, is the low technical ability of our laboratory staff; second is not using proper materials or techniques for this purpose; moreover, our pathologists had complained that the biopsy samples were too small for a proper PCR evaluation; although the general assumption is that PCR can detect the infection even in extremely limited volume samples. Ideally proton pump inhibitors should be discontinued before the endoscopy;[12,18] it was demonstrated that after 4 weeks of omeprazole treatment, the histological density of H. pylori in the antrum and corpus was reduced, while in the fundus was increased.[19] With a suspicion of high self medication among our patients, we hypothesize that drugs may cause an adverse impact on the accuracy of PCR test results.
Several previous studies have assessed the sensitivity and specificity of PCR methods to detect several primers in H. pylori’s gene loci. The results were very diverse in different reports. Lu et al[20] in their study comparing the power of five different PCR methods to detect H. pylori DNA in gastric tissues of patients with chronic gastritis, found that sensitivity of PCR methods is not satisfactory in detecting H pylori; they finally concluded that this observation can be related to sequence polymorphism in the specified loci. Smith et al[21] in their study of PCR methods for diagnosis of H. pylori infection in gastric tissues also reported a low sensitivity of 56% for glmM gene, but they found a sensitivity of 100% for 26kDa gene primer, but specificity of 44% for ureA. They also found that 68% of biopsies that showed positive amplification in all three genes were positive for the cagA gene while this proportion was just 43% in our survey. On the other hand, Lage et al[13] reported that there were no false positive or negative biopsies amplified by the glmM in their study. In this study, however, we reached to a sensitivity of 32% for 26kDa and specificity of 83% for ureA gene primer and comparable results for all the other gene primers investigated.
The high number of false negative results reported in this study, can not be solely interpreted by gene polymorphism; although, one may claim that the gold standard criteria used in this study for determination of positive and negative cases may not be of enough accuracy, our reason for using these criteria was that they were previously reported as high sensitivity and specificity methods and were also used as gold standard criteria for the same purpose by previous investigators.[22] PCR is a time consuming and expensive procedure with need for highly trained staff performing it. In the developing countries, the advantages of using PCR for detecting microorganisms such as H. pylori may be lost owing to the scenario of low healthcare funds aswell as apaucity of well-trained experts. Our study demonstrated that using PCR methods for detection of Helicobacter pylori does not have a high diagnostic accuracy rate.
References
1. Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175–86.
2. Ho SA, Hoyle JA, Lewis FA, Secker AD, Cross D, Mapstone NP, et al. Direct polymerase chain reaction test for detection of Helicobacter pylori in humans and animals. J Clin Microbiol. 1991;29:2543–9.
3. Mobley HL, Hu LT, Foxal PA. Helicobacter pylori urease:properties and role in pathogenesis. Scand J Gastroenterol Suppl. 1991;187:39–46.
4. Graham DY. Helicobacter pylori infection is the primary cause of gastric cancer. J Gastroenterol. 2000;35:90–7.
5. Li C, Ha T, Ferguson DA Jr, Chi DS, Zhao R, Patel NR, et al. A newly developed PCR assay of H. pylori in gastric biopsy, saliva, and feces. Evidence of high prevalence of H. pylori in saliva supports oral transmission. Dig Dis Sci. 1996;41:2142–9.
6. Pacheco N, Mago V, Gómez I, Gueneau P, Guelrud M, Reyes N, et al. Comparison of PCR and common clinical tests for the diagnosis of H. pylori in dyspeptic patients. Diagn Microbiol Infect Dis. 2001;39:207–10.
7. Price AB. The Sydney system: histological division. J Gastroenterol Hepatol. 1991;6:209–22.
8. Deltenre M, Glupczynski Y, De Prez C, Nyst JF, Burette A, Labbé M, et al. The reliability of urease tests, histology and culture in the diagnosis of Campylobacter pylori infection. Scand J Gastroenterol Suppl. 1989;160:19–24.
9. Marais A, Monteiro L, Occhialini M, Pina M, Lamouliatte H, Megraud F. Direct detection of Helicobacter pylori resistance to macrolides by a polymerase chain reaction/DNA enzyme immunoassay in gastric biopsy specimens. Gut. 1999;44:463–7.
10. Said RM, Cheah PL, Chin SC, Goh KL. Evaluation of a new biopsy urease test: Pronto Dry, for the diagnosis of Helicobacter pylori infection. Eur J Gastroenterol Hepatol. 2004;16:195–9.
11. Schnell GA, Schubert TT, Barnes WG, Rupani MK: Comparison of urease, H & E and culture tests for Helicobacter pylori. Gastroenterology (abstract). 1998;94:410.
12. Yakoob J, Jafri W, Abid S, Jafri N, Abbas Z, Hamid S, et al. Role of rapid urease test and histopathology in the diagnosis of Helicobacter pylori infection in a developing country. BMC Gastroenterol. 2005;5:38.
13. Lage AP, Godfroid E, Fauconnier A, Burette A, Butzler JP, Bollen A, et al. Diagnosis of Helicobacter pylori infection by PCR: comparison with other invasive techniques and detection of cagA gene in gastric biopsy specimens. J Clin Microbiol. 1995;33:2752–6.
14. Basso D, Navaglia F, Cassaro M, Scrigner M, Toma A, Dal Bo N, et al. Gastric juice polymerase chain reaction: an alternative to histology in the diagnosis of Helicobacter pylori infection. Helicobacter. 1996;1:159–64.
15. Lin SY, Jeng YS, Wang CK, Ko FT, Lin KY, Wang CS, et al. Polymerase chain reaction diagnosis of Helicobacter pylori in gastroduodenal diseases: comparison with culture and histopathological examinations. J Gastroenterol Hepatol. 1996;11:286–9.
16. Thijs JC, van Zwet AA, Thijs WJ, Oey HB, Karrenbeld A, Stellaard F, et al. Diagnostic tests for Helicobacter pylori: a prospective evaluation of their accuracy, without selecting a single test as the gold standard. Am J Gastroenterol. 1996;91:2125–9.
17. Roosendaal R, Kuipers EJ, van den Brule AJ, Peña AS, Uyterlinde AM, Walboomers JM et al. Importance of the fiberoptic endoscope cleaning procedure for detection of Helicobacter pylori in gastric biopsy specimens by PCR. J Clin Microbiol. 1994;32:1123–6.
18. Dickey W, Kenny BD, McConnell JB. Effect of proton pump inhibitors on the detection of Helicobacter pylori in gastric biopsies. Aliment Pharmacol Ther. 1996;10:289–93.
19. Logan RP, Walker MM, Misiewicz JJ, Gummett PA, Karim QN, Baron JH. Changes in the intragastric distribution of Helicobacter pylori during treatment with omeprazole. Gut. 1995;36:12–6.
20. Lu JJ, Perng CL, Shyu RY, Chen CH, Lou Q, Chong SKF, et al. Comparison of five PCR methods for detection of Helicobacter pylori DNA in gastric tissues. J Clin Microbiol. 1999;37:772–4.
21. Smith SI, Oyedeji KS, Arigbabu AO, Cantet F, Megraud F, Ojo OO, et al. Comparison of three PCR methods for detection of Helicobacter pylori DNA and detection of cagA gene in gastric biopsy specimens. World J Gastroenterol. 2004;10:1958–60.
22. Vinette KM, Gibney KM, Proujansky R, Fawcett PT. Comparison of PCR and clinical laboratory tests for diagnosing H. pylori infection in pediatric patients. BMC Microbiol. 2004;4:5.
|