K. Madan, 1 Y. Batra, 1 J.K. Jha, 1 Shakti Kumar, 1 Nancy Kalra, 1 S.B. Paul, 2 R. Singh, 3 S. Duttagupta, 4 S.K. Panda,4 , S.K. Acharya1
Department of Gastroenterology,1
Radiology,2 Biostatistics,3 and Pathology4
All India Institute of Medical Sciences
New Delhi - 110029
India
Corresponding Author:
Dr. SK Acharya
Email: subratacharya2004@yahoo.com
Abstract
Background: Hepatitis B virus (HBV) DNA detection and quantification are now playing an increasing role in the assessment of disease activity and response to therapy. However, viraemia levels which define various stages of HBV infection have not yet been established.
Aim: To define viraemia levels which describe various stages of chronic hepatitis B virus infection.
Methods: In a retrospective study, stored sera samples of chronic hepatitis B virus (CHB) infected patients registered at AIIMS liver clinic, from January 1996 to June 2005 were subjected to competitive, quantitative PCR analysis.
Results: The median HBV DNA load was lowest among carriers and highest among patients with chronic hepatitis B [0 (0-8) vs. 7 (0-12) log10 copies/ml, respectively; p<0.05]. As compared to chronic hepatitis patients the DNA load was also lower among cirrhotics [7 (0-12) vs. 4.5 (0-8) log 10 copies/ml, respectively; p<0.05] and hepatocellular cancer patients [ 7(0-12) vs. 0 (0-8) log10 copies/ml, respectively; p<0.05]. Patients with carriers had a DNA load which was significantly lower than e antigen negative CHB [0 (0-8) vs. 6 (0-10)log10 copies/ml; p<0.05] or e antigen positive CHB [0 (0-8) vs 8 (0-12) log10 copies/ ml; p<0.05]. A threshold of 3.5 log10 copies/ml had sensitivity and specificity of 83% and 58% respectively in differentiating carriers from e antigen negative CHB. There was a strong positive correlation of HBV DNA load with inflammatory grade (R=0.334; p=0.0001), fibrosis stage (R=0.276; p=0.001) and ALT levels (R=0.378; p=0.0001). 82% (9/11) of those who lost e antigen had a decline in HBV DNA levels to <5 log10 copies/ml, whereas only 12.5% (1/8) of those who did not lose e antigen had a decline in DNA load below this level.
Conclusions: HBV DNA viraemia levels correlate positively with the inflammatory grade, fibrosis stage and ALT levels. Most patients who loose e antigen have a decline in DNA load to below 5 log10copies/ml. Further prospective studies employing repeated measurements are required to define a threshold to differentiate between HBV carriers and e antigen negative CHB.
|
48uep6bbphidvals|225 48uep6bbphidcol2|ID 48uep6bbph|2000F98CTab_Articles|Fulltext Hepatitis B virus (HBV) infection is associated with significant morbidity and mortality due to end-stage liver disease and liver cancer. In India, it is a major public health problem, with an HBsAg carrier frequency of about 4% and estimated 40 million people infected at any given time.[1] The clinical spectrum of chronic hepatitis B virus infection ranges from silent histologically inactive carrier state to aggressive chronic hepatitis, cirrhosis and hepatocellular carcinoma (HCC). 15- 40% of patients with chronic HBV infection have been shown to progress to cirrhosis and end stage liver disease.[2]
Recent development of molecular biology techniques has shifted the focus of HBV diagnosis from serology to genome detection. HBV DNA detection and quantification are now playing an increasing role in the assessment of viral activity and response to therapy. It has been demonstrated in previous studies, that HBV DNA levels are usually in excess of 10[5] among patients of CHB who are e antigen positive.[3,4,5] Chronic hepatitis B patients who are e-antigen negative have a lower DNA load as compared to e antigen positive patients, but their loads are higher than those of inactive carriers. A cut-off level of 100,000 copies/ml of serum has been recommended to differentiate between carriers and patients with chronic hepatitis.[6,7] However this has not been validated and a recent study from Greece has suggested that using a cut-off > 30,000 copies/ml has a higher sensitivity in correctly differentiating e antigen negative chronic hepatitis B from HBV carriers.[8] But even a cut-off of 30,000 copies/ml misclassified 7% of inactive carriers and 30% of patients with HBeAg-negative chronic hepatitis where testing was done only once.[3] But most previous investigators defined HBV carriers based on persistently normal ALT levels without confirming the absence of disease on liver biopsy. Further, level of HBV viraemia in different clinical forms of HBV infection is not known. Another lacuna in the existing knowledge is the inconsistent correlation between HBV viraemia levels and histology or ALT.
Thus we planned to carry out this study with the following aims, 1) to define the HBV viraemia levels among patients with different clinical forms of HBV infection, and 2) to find out a threshold value of HBV DNA load which could differentiate patients who have diseased livers from those who are only carrying the virus. This is important in order to identify that group of HBV infected patients who are more likely to progress to advanced stages of the disease.
Methods
Study design
This was a retrospective study in which we analysed stored sera of patients with chronic hepatitis B virus infection, collected during their visit to the Liver Clinic of AIIMS between January 1996 to June 2005.
Patients
The study group patients were regularly followed up at the Liver Clinic at 1 to 6 monthly intervals as indicated. During each visit, the patients underwent routine biochemical tests as detailed later. Viral serologies including hepatitis B surface antigen (HBsAg), hepatitis B e antigen (HBeAg) and antibodies to hepatitis B e antigen (anti-HBeAg) were also performed at an interval of 3 months for patients receiving some form of therapy or 6 –12 monthly for patients not receiving any therapy. The sera samples were stored at -70°.
Serologic assays
The serum samples were investigated for HBV infection, using commercial micro-enzyme-linked immunosorbent assay (ELISA) for HBsAg, HBeAg , anti-HBe, antibodies to hepatitis B core antigen (anti-HBcAg) (Organon Teknika, Boxtel, RM, The Netherlands) and antibodies to hepatitis C virus (anti-HCV) (Xcyton, Bangalore, India). The tests were performed according to the manufacturer’s instructions.
HBV DNA load measurements
HBV DNA load was measured using the competitive PCR developed at our own lab. A PCR reaction was set up with HBV core primers PS4 and PA2 as qualitative detection PCR. The only difference being that, instead of a 5 µl template, 2.5 ml of unknown sample DNA and 2.5µl of internal standard at known dilutions were added. Each unknown sample to be quantitated was titrated against 3 log dilutions at internal standard-10[4], 10[6], 10[8] copies/ml. The rest of the outer and nested PCR was the same as has been described in a qualitative analysis published previously.[9] From the post amplified product 15µl of PCR reaction mixture was taken in a separate tube and digested with Hind III enzyme (Amersham, Pharmacia, UK). The PCR product was digested with Hind III and kept for 8-10 hours at 37°C for complete digestion. The digested PCR product was dissolved in 2% agarose gel containing 0.05% ethidiium bromide. This should ideally show three bands as 634 bp for sample DNA (amplification product), and 392 bp and 242 bp for standard DNA on digestion with Hind III (Figure 1). Unknown samples can be quantified by densitometric analysis and gel blot software at known standard. The intensity of the ethidium fluorescence associated with the DNA band is proportional to the amount of DNA.
The relative ratio of the OD value of 392 bp to the OD of 634 bp band was plotted against the copy numbers of the competitor (mutant) HBV DNA. The copy number of HBV DNAper reaction were determined by calculating the ratio of absorbance of the unknown standard.
2.5 µl of 30 µl of HBV DNA elute was taken as a template which was isolated from 100 µl of serum. For each sample the viral load was calculated by dividing HBV DNA copies calculated densitometrically, by 30 and multiplying by 1000 to obtain the approximate number of viral DNA copies/ml of serum. The values of DNA load were expressed as log[10] copies/ml of serum.
This method of DNA quantification has shown good correlation with the results obtained by real time PCR method.
Liver biopsy
Liver biopsy was performed in a select set of patients who gave their full and voluntary consent after the nature of the disease and the particulars of the procedure were explained in detail. The liver biopsy was performed ercutaneously, using a 16 G Menghini’s aspiration needle under lignocaine local anaesthesia. The liver biopsy specimen was fixed in 10% formalin and sections were stained with haematoxylineosin, reticulin, and masson’s trichrome stains. Immunohistochemical staining was also done to detect the presence of HBsAg and HBcAg in the liver tissue, using monoclonal antibodies (Dakopats, Copenhagen, Denmark). The antigens were detected by the streptavidin-biotin complex technique with 3,3’ diaminobenzidine tetrahydrochloride (D- 0426; Sigma, St. Louis, MO) as chromogen. Brown colouration indicated positive staining. Histological grading and staging was done according to the scheme given by Ishak et al.[10]
Diagnostic criterias
Carriers: Carriers were defined as those patients with hepatitis B infection who on liver biopsy had no evidence of liver injury (histological activity index<4 and stage<2) or in those in whom histology was not available, by presence of persistently normal ALT for at least 6 months.
Chronic hepatitis B: HBsAg positive patients who on liver biopsy had evidence of liver injury as defined by an HAI > 4 and/or fibrosis stage > 2, irrespective of the level of ALT were labelled as chronic hepatitis B patients. Patients were classified as e Ag positive or negative by the presence or absence of this antigen respectively.
Reactivation of chronic hepatitis B: Patients who presented with positive HBsAg in the presence of raised ALT above 5 times upper limit of normal with or without clinical jaundice and in whom IgM antibodies to hepatitis B core antigen were not detectable were diagnosed as reactivation of chronic hepatitis B.
Hepatitis B related cirrhosis: Patients who had clinical, radiological, endoscopic or histological evidence of cirrhosis of liver with HBsAg positivity in the serum along with absence of history or markers pointing to an alternative cause of liver damage, were diagnosed as HBV related cirrhosis.
Hepatocellular carcinoma (HCC): HCC due to hepatitis B was diagnosed when in a patient, there was radiological evidence of a mass lesion which showed arterial enhancement in the triphasic contrast enhanced CT scan and in MR contrast enhanced scan of the liver or raised alfafeto protein > 300 ng / ml with arterial enhancement of the mass lesion in one of the forementioned imaging modalities or fine needle aspiration from the mass showing characteristics of hepatocellular cancer with a positive HBsAg or positive anti bodies to HBc Ag in the absence of any other risk factor for the development of HCC.
Statistical analysis
Quantitative variables were expressed as median (range). Qualitative variables were expressed as frequencies. For comparison of quantitative data among the groups, the Kruskal Wallis test was used. Post hoc analysis was done using the Newman Kaulli’s test. For discrete variables, the chi-square test was used. Spearman correlation was used to demonstrate correlation between DNA load and histological severity and ALT levels. A Receiver operating characteristics (ROC) curve analysis was done in order to find a cut-off value which could differentiate HBV carriers from e Ag negative chronic hepatitis B patients. A two sided p value of less than 0.05 was considered significant.
Ethical clearance was taken from the Institute’s Ethics Committee and informed consent was taken from all patients for the use of stored sera; all patients were on follow up or could be contacted via the telephone.
Results
Over a ten and a half year period, 470 cases of hepatitis B related chronic liver disease were registered at the Liver Clinic of the All India Institute of Medical Sciences. Of these 85, 170, 15, 97 and 103 patients were classified as HBV carriers, chronic hepatitis B, reactivation of chronic hepatitis B, HBV related cirrhosis and HBV related HCC, respectively. The baseline characteristics of the 5 groups are given in Tablel
DNA load in the 5 groups (Figure 2)
Baseline DNA loads were available in 193 patients, of which 47 were carriers, 85 were patients with chronic hepatitis B, 7 had reactivation of CHB, 30 had cirrhosis and 24 had HCC. The median DNA load was significantly lower among carriers as compared to patients with chronic hepatitis (with 0 (0-8) vs. 7 (0-12) log[10] copies/ml; p<0.05] or without reactivation [0 (0-8) vs. 7 (0-10) log[10] copies/ml]. Chronic hepatitis patients had a significantly higher DNA load than patients who had chronic hepatitis with reactivation [7 (0-12) vs. 7 (0-10) log[10] copies/ml; p<0.05], cirrhosis [7 (0-12) vs. 4.5 (0-8) log[10] copies/ml; p<0.05] or HCC [7 (0-12) vs. 0 (0-8) log[10] copies/ ml; p<0.05].
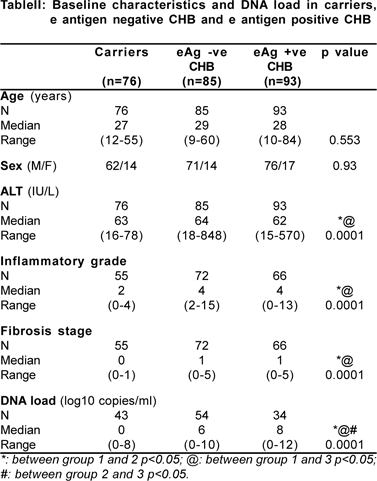
DNA load in carriers, e Ag positive and e Ag negative chronic hepatitis B
The baseline characteristics of these three groups along with the respective DNA load are depicted in Table II. The median DNA load was lowest in patients who were carriers when compared to e antigen negative CHB [0 (0-8) vs. 6 (0-10) log[10] copies/ml; p=0.0001] and e antigen positive CHB [0 (0- 8) vs. 8 (0-12) log[10] copies/ml; p=0.0001]. The proportion of patients with undetectable viraemia was significantly higher in carriers [55.8% (24/43)], as compared to e negative CHB [16.7% (9/54) and e positive CHB [9% (6/34)]. 26% of our carriers had a high DNA load of more than 5 log10 copies/ml (according to the NIH definition of carriers). Thus we had carriers with low and high DNA load. It would be interesting to prospectively evaluate any difference in the natural courses of carriers with low and high viral load respectively.
We also plotted an ROC curve in order to find out the threshold level of HBV viraemia which could correctly differentiate between HBV carriers and e antigen negative chronic hepatitis patients (Figure 3). The area under the curve was 0.755 (p=0.0001; CI:0.65-0.85) and a cut off of 3.5 log[10] copies/ml had good sensitivity of 83.1% but poor specificity of 58.1% in identifying patients who had e antigen negative chronic hepatitis B.
Correlation of DNA load with histology and ALT levels
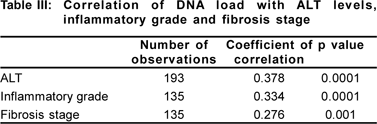
Liver biopsy findings were available in 236 patients, of which, 61, 129, 13, 28 and 5 were carrier, CHB , CHB with reactivation, cirrhosis and HCC patients, respectively. Spearman’s correlation was used to test for correlation between DNA load and histological grade, histological stage and ALT levels. There was significant positive correlation of DNA load and histological grade (R=0.334; p=0.0001), stage of fibrosis (R=0.276; p=0.001) and ALT levels (R=0.378; p=0.0001) (Table III) (Figures 4a, 4b, 4c).
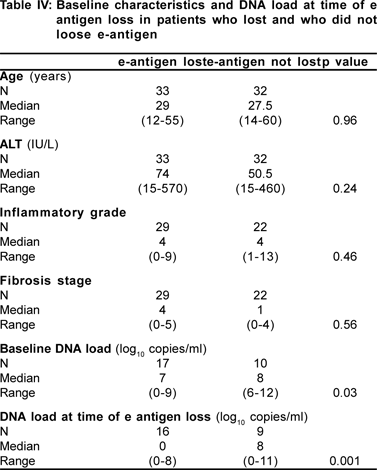
Threshold DNA load for conversion to e antigen negative state
We wanted to find out whether there was any threshold DNA level at which there occurred HBeAg seroconversion, either spontaneously or therapy induced. However due to the retrospective nature of this study, there were not enough observations available for performing this analysis. But we analysed the DNA load available in the groups which lost e antigen versus those who did not lose e antigen. The baseline characteristics of the group which lost and the one which did not lose the e antigen were similar except for a higher baseline DNA load in the latter. The median duration of therapy before HBe Ag was lost in 11 patients (who had both baseline DNA value and DNA value at time of e antigen loss) was 4 (2.5-23) months. There was a significant reduction in mean (4.8 to 2.3 log[10]copies/ml; p=0.02) and median (7log[10 ]to undetectable; p=0.02) DNA load at the time the e antigen became undetectable. 82% (9/11) of those who lost the e antigen had a decline in the HBV DNA level to less than 5 log[10]copies/ml, however only 12.5% (1/8) of those who did not lose the e antigen had a decline in DNA load below this level (p=0.005). However on plotting an ROC curve, no threshold value of DNA level was found which could define conversion of e antigen positive to the e antigen negative state (Table IV).
Discussion
Based on the relative interaction between host immune response and viral factors, the natural course of hepatitis B infection can be divided in to fours phases. These are the immunotolerant phase, immune clearance phase, nonreplicative phase, and phase of resolution.[11] These usually correspond to different clinical forms of chronic hepatitis B infection. In the present study we evaluated HBV DNA loads in various clinical forms of chronic hepatitis B infection. HBV DNA levels were found to be significantly lower in carriers of hepatitis B as compared to patients with chronic hepatitis. The highest levels were found in patients who had chronic hepatitis. Patients in the inactive carrier state are said to be in the third phase (non-replicative phase) of the natural course of infection with very low levels of viraemia, undetectable by non-PCR based assays. We found that 56% of our carriers did not have detectable viraemia even though we used a PCR based assay, the detection limit of which is 1000 HBV DNA copies/ml. The highest level of viraemia was found in patients who had chronic hepatitis. During the phase of immune clearance, the immune system of the host starts clearing up the virus infected hepatocytes resulting in liver injury as well as start of decline of viraemia which may persist for a very long time and during severe flares, may either result in clearance of virus or further aggravation of injury. During severe flares or clinical reactivation, the virus load would understandably drop further due to severe immune clearance as was found in our patients with reactivation, in whom the virus load was significantly lower than patients with chronic hepatitis without reactivation. In patients with hepatocellular carcinoma, at the time of diagnosis, the HBV DNA levels were significantly lower than in patients with chronic hepatitis. It is known that in patients with HCC the HBV genome may be incorporated in the genome of the host cells and would not be circulating in the serum.
As has been demonstrated in other studies, the DNA levels were highest among patients who had e antigen positive chronic hepatitis B, followed by patients who had e antigen negative chronic hepatitis B and the lowest levels were found among carriers.[3,12,13,14]
In the present study, although the DNA levels in patients with e antigen positive CHB were higher than in patients with e antigen negative CHB, the difference was not significant.
One important issue is to find a level of viraemia that can differentiate between HBV carriers (inactive state) and e antigen negative chronic hepatitis B (diseased state). We tried to determine this threshold level by plotting an ROC curve taking the DNA load of inactive carriers of e antigen negative CHB patients. Taking a cut off of 3.5 log[10] copies/ml, the sensitivity and specificity were 83 and 58%, respectively. The National Institutes of Health workshop on management of hepatitis B did suggest a cut off of 10[5] copies /ml, this was an arbitrary value. [6,7] Subsequent studies have tried to validate this cut off value, but it has been seen that taking this level of viraemia as the cut off, 45% of e antigen negative CHB patients would be misclassified.[3] Manesis et al suggested that a cut off value of 3 X 10[3] would be a better cut off for differentiating these two groups, but even this level was shown to misclassify 7% of inactive carriers and 30% of patients with HBeAg negative CHB if testing was only done at presentation.3,8 Thus in accordance with the results of Chu et al, we found that no single level of HBV DNA load reliably differentiates these two groups. Further, repeated measurements are more reliable than a single baseline value. In our study repeated measurements were not available for many patients. However we are in the process of analysing stored sera as well as planning a prospective study in order to answer these questions. Even in a prospective study, getting repeated measurements of HBV DNA levels in patients with e antigen negative CHB would be difficult, as most of them would have been put on some form of antiviral therapy. One major strength of our study over previous studies is the definition of inactive carriers on the basis of absence of disease on histology. Previous studies have defined carrier state based on the presence of persistently normal ALT levels over a variable time period from 6 to 24 months. It is known that even patients who have normal ALT levels may have significant histological disease.[15,16,17]
Therefore it is possible that the previous studies could have included CHB patients in their group of carriers resulting in higher DNA levels in some of the patients included in the carrier group Since hepatitis B is not a cytopathic virus, it is not surprising that most studies have not been able to demonstrate any correlation between HBV DNA load and histological severity of disease, because active histological disease would mean immune mediated clearance of virally infected hepatocytes and lower viraemia levels. Thus when histological injury is severe the level of viraemia is expected to be low. These studies mainly observed the correlation between HBV DNA levels and histology among e antigen positive and e antigen negative CHB patients and found positive correlation only in patients with e antigen negative CHB.[4,14,17] However, in the present study we have included all clinical forms of hepatitis B infection, including cirrhosis and HCC and have demonstrated that there is positive correlation between HBV DNA levels and the inflammatory grade, fibrosis stage and ALT levels.
Another unanswered question in the management of chronic hepatitis B is the DNA level which defines e antigen loss or e antigen seroconversion (spontaneous or therapyinduced). In one study, among 23 patients who received lamivudine, 6 of 12 patients whose serum HBV DNA levels reduced to less than 4 log[10]copies /ml, developed HBe antigen seroconversion, whereas none of the 11 patients whose serum HBV DNA levels remained above 4 log[10]copies/ml did so.[18] In the present study the HBV DNA load declined by a mean of at least 2 log[10]copies/ml at the time of e antigen loss. Significantly higher number of patients had a decline in HBV DNA levels to < 5 log[10]copies/ml at the time of e antigen loss as compared to those who did not lose e antigen (82% versus12.5%). But we could not find a threshold level that could define e antigen loss. This may be related to the small number of observations in each group (11 and 8 only) Similar to our data, Chu et al also demonstrated a fall in HBV DNA levels by at least 3 log[10]copies/ml at the time of e antigen loss, but could not detect a single value which could differentiate the two groups.[3]
The lacunae in the present study are the retrospective nature of the study and the fact that we have only one time DNA load measurements in HBV carriers and untreated CHB patients. It is well known that the HBV DNA loads may fluctuate widely with time, especially among e antigen negative CHB patients. [19,20]
To conclude, we have successfully demonstrated that the highest levels of HBV viraemia are detected in e antigen positive patients and lowest levels occur in carriers and HCC patients. It is difficult to define a single DNA value, which could differentiate between carriers and e negative CHB patients, although a value of 3.5 log[10] copies/ml provides a sensitive cut-off even though the specificity is low. There is significant positive correlation of HBV DNA levels with histological severity of disease and with ALT levels. Although there is a significant fall in HBV viraemia at the time of e antigen loss, no single value of HBV DNA load can reliably predict this outcome. Prospective studies, using repeated DNA load measurements may be able to answer these questions.
Reference
1. Tandon BN, Acharya SK, Tandon A. Epidemiology of hepatitis B virus infection in India. Gut. 1996;38:S56-9.
2. Liaw YF, Tai DI, Chu CM, Chen TJ. The development of cirrhosis in patients with chronic type B hepatitis: a prospective study. Hepatology. 1988;8:493–6.
3. Chu CJ, Hussain M, Lok ASF. Quantitative serum HBV DNA levels during different stages of chronic hepatitis B infection. Hepatology. 2002;36:1408–15.
4. Lindh M, Horal P, Dhillon AP, Norkrans G. Hepatitis B virus DNA levels, pre-core mutations, genotypes and histological activity in chronic hepatitis B. J Viral Hepatitis. 2000;7:258–67.
5. Niitsuma H, Ishii M, Miura M, Kobayashi K, Toyota T. Low level hepatitis B viremia detected by polymerase chain reaction acoompanies the absence of HBe antigenenmia and hepatitis in hepatitis B virus carriers. Am J Gastroenterol. 1997;92:119–23.
6. Lok AS, McMahon BJ; Practice Guidelines Committee, American Association for the Study of Liver Diseases. Chronic hepatitis B. Hepatology. 2001;34:1225–41.
7. Lok AS, Heathcote EJ, Hoofnagle JH. Management of hepatitis B: 2000-summary of a workshop. Gastroenterology. 2001;120:1828–53.
8. Manesis EK, Papatheodoridis GV, Sevastianos V, Cholongitas E, Papaioannou C, Hadziyannis SJ. Significance of hepatitis B viremia levels determined by a quantitative polymerase chain reaction assay in patients with hepatitis B e antigen-negative chronic hepatitis B virus infection. Am J Gastroenterol. 2003;98:2261–7.
9. Chaudhuri V, Tayal R, Nayak B, Acharya SK, Panda SK. Occult hepatitis B virus infection in chronic liver disease: Full length genome and analysis of mutant surface promoter. Gastroenterology. 2004;127:1356–71.
10. Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudal F, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696–9.
11. Ganem D, Prince AM. Hepatitis B e-antigen negative chronic hepatitis B. Hepatology 2001;34:617–24.
12. Kessler HH, Preininger S, Stelzl E, Daghofer E, Santner BI, Marth E, et al. Identification of different stages of hepatitis B virus infection with a quantitative PCR assay. Clin Diagn Lab Immun. 2000;7:298–300.
13. Fujiwara K, Yokosuka O, Ehata T, Chuang WL, Imazeki F, Saisho H, et al. The two different stages of hepatitis B virus DNA in asymptomatic carriers: HBeAg positive versus anti-HBe positive asymptomatic carriers. Dig Dis Sci. 1998;43:368–76.
14. Yuen MF, Ng IOL, Fan ST, Yuan HJ, Wong DKH, Yuen JCH, et al. Significance of HBV DNA levels in liver histology of HBe Ag and anti-HBe positive patients with chronic hepatitis B. Am J Gastroenterol. 2004;99:2032–7.
15. Brunetto MR, Cerenzia MT, Oliveri F, Piantino P, Randone A, Calvo PL, et al. Monitoring the natural course and response to therapy of chronic hepatitis B with an automated semi-quantitative assay for IgM anti-HBc. J Hepatol. 1993;19:431–6.
16. Mels GC, Bellati G, Leandro G, Brunetto MR, Vicari O, Borzio M, et al. Fluctuations in viremia, aminotransferases and IgM antibody to hepatitis B core antigen in chronic hepatitis B patients with disease exacerbations. Liver 1994;14:175–181.
17. Zavaglia C, Mondazzi L, Maggi G, Iamoni G, Gelosa F, Bellati G, et al. Are alanine aminotransferase, hepatitis B virus DNA or IgM antibody to hepatitis B core antigen serum levels predictors of histological grading in chronic hepatitis B? Liver. 1997;17:83–7.
18. Gauthier J, Bourne EJ, Lutz MW, Crwother LM, Dienstag JL, Brown NA, et al. Quantitation of hepatitis B viremia and emergence of YMDD variants in patients with chronic hepatitis B treated with lamivudine. J Infect Dis. 1999;180:1757–62.
19. Hadziyannis S, Bramou A, Alexopoulou A, et al. Immunopathogenesis and natural course of anti-HBe-positive chronic hepatitis with replicating B virus. In: Hollinger FB. Lemon SM, Margolis HS, eds. Viral hepatitis and liver disease. Baltimore: Williams and Wilkins, 1991:673–6.
20. Noborg U, Gusdal A, Horal P, Lindh M. Levels of viremia in subjects with serological markers of past or chronic hepatitis B virus infection. Scan J Infect Dis. 2000;32:249–52.
|